Trending Now
- 830 voters names go missing in Kavundampalayam constituency
- If BJP comes to power we shall consider bringing back electoral bonds: Nirmala Sitaraman
- Monitoring at check posts between Kerala and TN intensified as bird flu gets virulent in Kerala
Tamilnadu News
Battling the spread and development of the Corona virus
![]() October 14, 2021
October 14, 2021
The corona virus infection (COVID-19) is a global pandemic which is rapidly spreading around the world. WHO, Scientific experts, the government and volunteers are working together to track the viral variants and alerting countries to stop the spread. Corona virus causes miserable lung conditions associated with severe fever, cough, chest pain, diarrhea and shortness of breath. The virus mutates over time and adapts to different viral properties to enhance the spread. These viral variants are denoted as Alpha (α), Beta (β), Gamma (γ), and Delta (δ) increase the transmissibility and fatality with a decreasing effectiveness of the vaccine. Currently, several countries are at the beginning stage of the third wave, and there are speculations about subsequent fourth and fifth waves.
Taking this into consideration, Prof. J. Jeyakanthan, Head of the Department of Bioinformatics, Alagappa University, and his team of scholars have been engaged in continuous research in finding novel drugs against the corona virus for the past one and a half years.
Additionally, research is being carried out on computer-assisted detection of future mutations in Delta (δ) type corona virus, which is rapidly spreading in India.
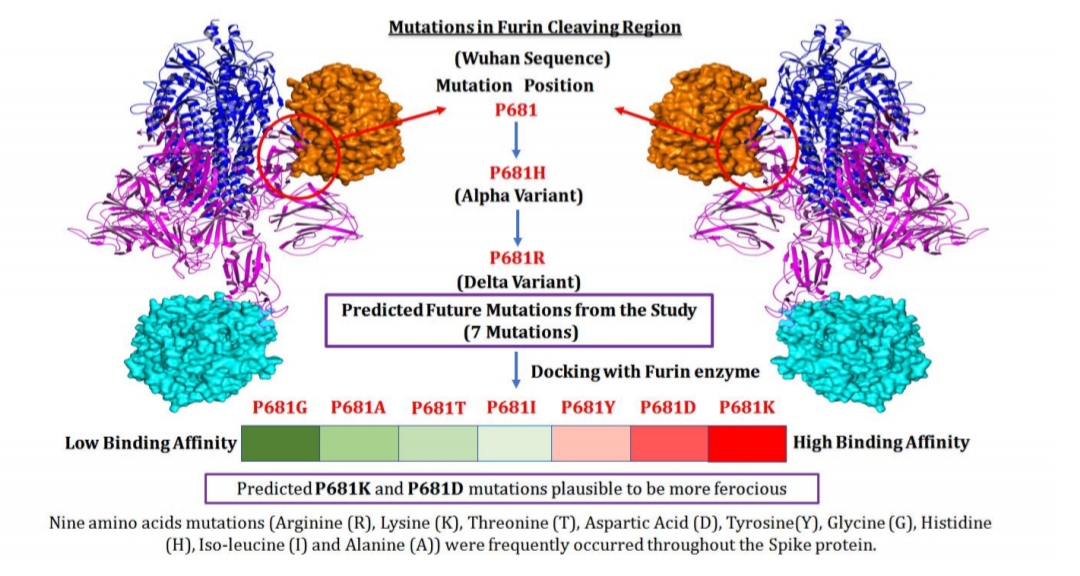
Spike protein (S-Protein: 1273 amino acids) of SARS-CoV-2 plays a key role in host receptor binding recognition and cell membrane fusion to
release the viral RNA material inside the human cell. Spike protein consists of two domains (S1 and S2 Subunits), and these have two cleavage sites (S1 and S2). Furin, Cathepsin, and TMPRSS2 are human proteins involves in proteolytic (cleavage) activity on S1 and S2 site for viral invasion. S1 subunit of SARS-CoV-2 consists of a Receptor Binding Domain (RBD) that binds with the host receptor hACE2 (Human Protein) while, S2 subunit enhances the ability of the virus fusion in the membrane for indispensable function (Figure 1). Center for Disease Control and Prevention (CDC) agency have reported the mutations in the Spike protein and classified them based on the impact of the variant. Thirteen types of Alpha (α), Ten types of Beta (β), Eleven types of Gamma (γ), and Fifteen types of Delta (δ) mutations have been reported for spike protein. Furthermore, genome sequencing and bioinformatics approaches can be used to detect the emergence of new mutations and evolutionary similarities rapidly.
Figure 1: Schematic representation of spike protein structure and cleavage region.
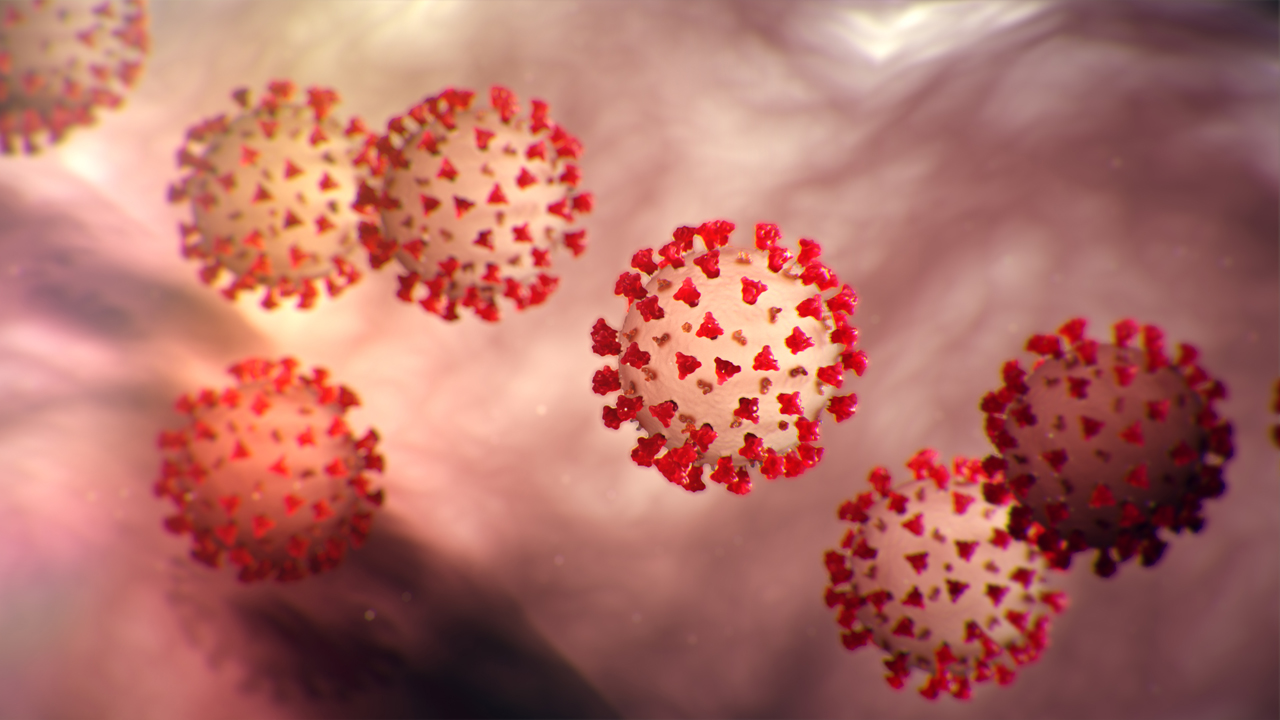
The spike protein sequences submitted from various research laboratories were downloaded from the GISAID database. As of now, approximately 4,208,359 hCoV-19 data have been reported and nearly 47,000 Indian spike proteins (spike protein sequences) were taken for further analysis. By keeping the Wuhan sequence as template, the spike protein sequences (Spike) in states such as Tamil Nadu (191), Andhra Pradesh (461), Telangana (494), Kerala (60), Karnataka (181), Delhi (545), Maharashtra (292), and Gujarat (501) were taken for this study. Based on the analysis, the amino acid mutations such as L18F, T19R, I68/H69/V70del, D80A, D138Y, G142D, 144del, E156G, F157del, R158G, D215G, A222V, L241/L242del, T478K, E484KQ, N501Y, A570D, D614G, H655Y, P681HR, A701V, T716I, D950N, S982A, and D1118H were observed. We found that people carry an enormous amount of Delta variant with P681R, which denotes that the Delta variant has a dominant spread around the country. P618R was known to be a driving force for the quick spread of Delta variants. Reports say that the mutation occurs at the Furin Cleavage region (S1 / S2) in the spike protein, which could increase the rate of infection and its severity. Computer assisted amino acids
mutations in the Furin Cleavage Region showed different binding affinity profile for the complex formation [Proline-681-Glycine (P681G), Proline-681-Alanine (P681A), Proline-681-Threonine (P681T), Proline-681-Isoleucine (P681I), Proline-681-Tyrosine (P681Y), Proline-681-Aspartic acid (P681D) and Proline-681-Lysine (P681K)].
Figure 2: Description of spike protein activity and subsequent changes found in this study
Based on this study, we highlighted that mutations, Proline-681-Lysine (P681K) and Proline-681-Aspartic acid (P681D) have greater binding affinity towards the Furin protein (Figure 2). If these predicted mutations arise in Corona virus,it may cause more virulent infections and detrimental effects in future. Consistently, our research team is working on determining the structure of Covid-19 proteins through.crystallography and computational biology approaches. Even as efforts on to find effective drugs or vaccines, curbing the spread of Covid-19 starts with every individual, by following social distancing, wearing a proper mask and getting vaccinated.